Classification Intuition#
[2]:
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from mpl_toolkits import mplot3d
from graphpkg.static import plot_classification_boundary
import tensorflow as tf
from sklearn.datasets import make_blobs, make_classification, make_regression
import warnings
warnings.filterwarnings("ignore")
Basic Classification#
Target here to generate a data that has only two classes and it can be classified by a linear model also. Like a linear hyperplane can also be a good decision boundary.
[2]:
X, y = make_classification(n_samples=500, n_features=2, random_state=30, \
n_informative=1, n_classes=2, n_clusters_per_class=1, \
n_repeated=0, n_redundant=0)
print(X.shape, y.shape)
df = pd.DataFrame(X,columns=['x1','x2'])
df['y'] = y
sns.scatterplot(data=df,x='x1',y='x2',hue='y')
(500, 2) (500,)
[2]:
<AxesSubplot:xlabel='x1', ylabel='x2'>
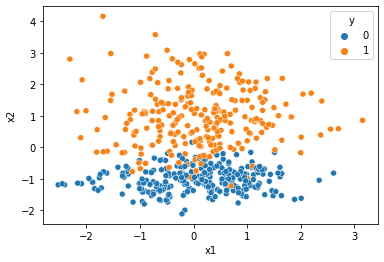
This is prety much it. linearly classifiable data with 2 classes.
Logistic Regression with Neural network (sigmoid)#
logistic regression is like a linear classification model.
- :nbsphinx-math:`begin{align}
y &= g(h(x))\ h(x) &= w x + b\ text{sigmoid } g(x) &= frac{1}{1 + e^{-x}}
end{align}`
So, actually after a linear weight and bias model, we need a sigmoid. It is actually called a perceptron.
[3]:
model = tf.keras.Sequential([
tf.keras.layers.Dense(units=1, activation='sigmoid')
])
model.compile(
optimizer=tf.optimizers.SGD(learning_rate=0.09),
loss=tf.keras.losses.BinaryCrossentropy(),
metrics=tf.keras.metrics.BinaryAccuracy()
)
history = model.fit(
df[['x1','x2']],
df.y,
epochs=500,
batch_size=32,
verbose=0,
validation_split = 0.3)
history_metrics = pd.DataFrame(history.history)
history_metrics['epochs'] = history.epoch
[4]:
fig,ax = plt.subplots(1,2,figsize=(10,5))
history_metrics.plot(x='epochs',y=['loss','val_loss'],ax=ax[0])
history_metrics.plot(x='epochs',y=['binary_accuracy','val_binary_accuracy'],ax=ax[1])
[4]:
<AxesSubplot:xlabel='epochs'>
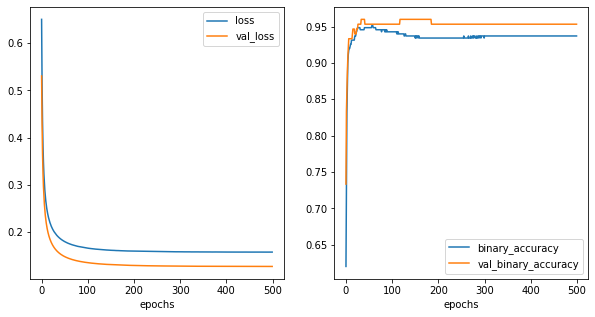
[5]:
y_hat = model.predict(df[['x1','x2']].values)
df['y_hat'] = np.array(y_hat>0.5,dtype='int')
fig,ax = plt.subplots(1,2,figsize=(10,5))
sns.scatterplot(data=df,x='x1',y='x2',hue='y',ax=ax[0])
sns.scatterplot(data=df,x='x1',y='x2',hue='y_hat', ax=ax[1])
[5]:
<AxesSubplot:xlabel='x1', ylabel='x2'>
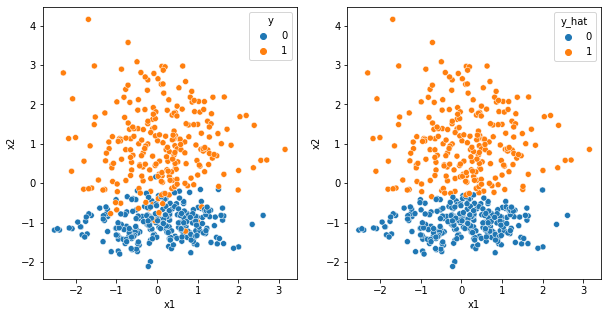
[6]:
tf.keras.utils.plot_model(model,show_layer_activations=True)
[6]:
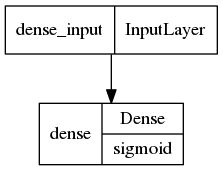
[7]:
plot_classification_boundary(model.predict,data=df[['x1','x2','y']].values,size=4,figsize=(7,5))
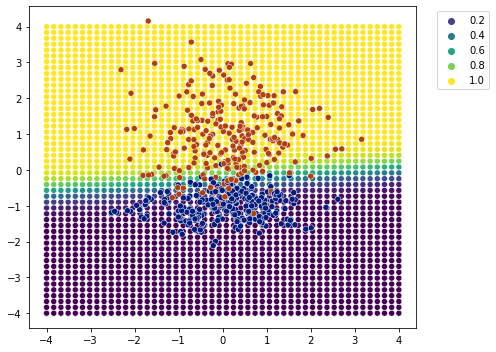
Looks to me that it worked out.
2 sigmoid layers#
[8]:
x1, x2 = make_regression(n_features=1,noise=10, random_state=0, n_samples=500)
x2 = (x2/100)**2
[9]:
df1 = pd.DataFrame()
df1['x1'] = x1[...,-1]
df1['x2'] = x2
df1['y'] = 0
df2 = pd.DataFrame()
df2['x1'] = x1[...,-1]
df2['x2'] = x2 + 0.5
df2['y'] = 1
df = pd.concat((df1,df2)).sample(frac=1)
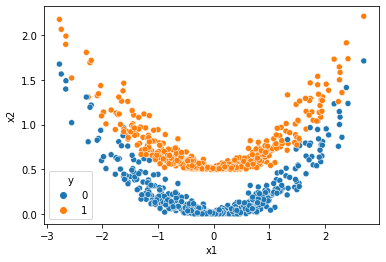
sns.scatterplot(data=df,x='x1',y='x2',hue='y')
[9]:
<AxesSubplot:xlabel='x1', ylabel='x2'>
[10]:
model = tf.keras.Sequential([
tf.keras.layers.Dense(units=10, activation='sigmoid'),
tf.keras.layers.Dense(units=1, activation='sigmoid')
])
model.compile(
optimizer=tf.optimizers.SGD(learning_rate=0.1),
loss=tf.keras.losses.BinaryCrossentropy(),
metrics=tf.keras.metrics.BinaryAccuracy()
)
history = model.fit(
df[['x1','x2']],
df.y,
epochs=500,
batch_size=32,
verbose=0,
validation_split = 0.3)
history_metrics = pd.DataFrame(history.history)
history_metrics['epochs'] = history.epoch
[11]:
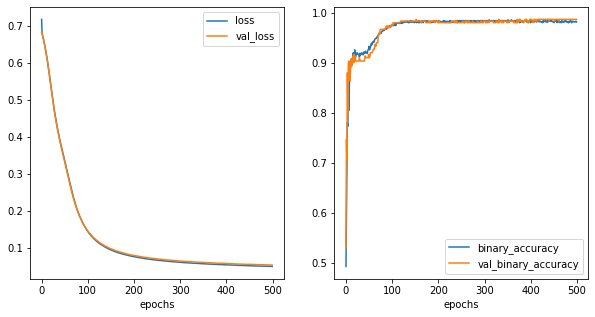
fig,ax = plt.subplots(1,2,figsize=(10,5))
history_metrics.plot(x='epochs',y=['loss','val_loss'], ax=ax[0])
history_metrics.plot(x='epochs',y=['binary_accuracy','val_binary_accuracy'], ax=ax[1])
[11]:
<AxesSubplot:xlabel='epochs'>
[12]:
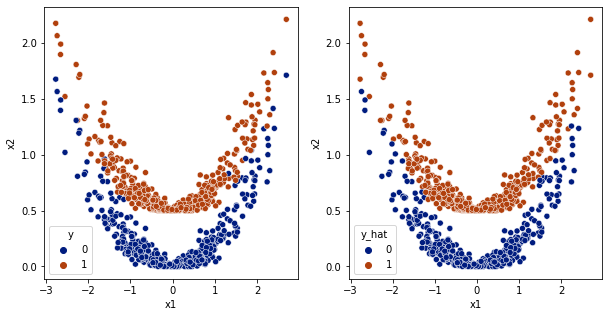
y_hat = model.predict(df[['x1','x2']].values)
df['y_hat'] = np.array(y_hat>0.5,dtype='int')
fig,ax = plt.subplots(1,2,figsize=(10,5))
sns.scatterplot(data=df,x='x1',y='x2',hue='y',ax=ax[0], palette='dark')
sns.scatterplot(data=df,x='x1',y='x2',hue='y_hat', ax=ax[1], palette='dark')
[12]:
<AxesSubplot:xlabel='x1', ylabel='x2'>
[13]:
tf.keras.utils.plot_model(model,show_layer_activations=True)
[13]:
[14]:
plot_classification_boundary(model.predict,data=df[['x1','x2','y']].values,size=3,\
figsize=(7,5), bound_details=100)
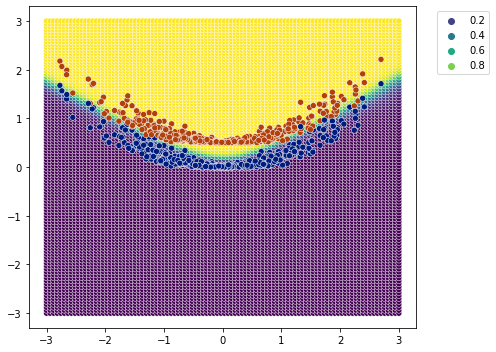
3 sigmoid layers#
[15]:
model = tf.keras.Sequential([
tf.keras.layers.Dense(units=10, activation='sigmoid'),
tf.keras.layers.Dense(units=10, activation='sigmoid'),
tf.keras.layers.Dense(units=1, activation='sigmoid')
])
model.compile(
optimizer=tf.optimizers.SGD(learning_rate=0.1),
loss=tf.keras.losses.BinaryCrossentropy(),
metrics=tf.keras.metrics.BinaryAccuracy()
)
history = model.fit(
df[['x1','x2']],
df.y,
epochs=500,
batch_size=32,
verbose=0,
validation_split = 0.3)
history_metrics = pd.DataFrame(history.history)
history_metrics['epochs'] = history.epoch
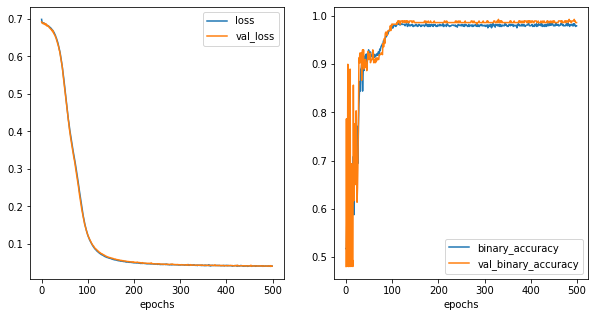
[16]:
fig,ax = plt.subplots(1,2,figsize=(10,5))
history_metrics.plot(x='epochs',y=['loss','val_loss'], ax=ax[0])
history_metrics.plot(x='epochs',y=['binary_accuracy','val_binary_accuracy'], ax=ax[1])
[16]:
<AxesSubplot:xlabel='epochs'>
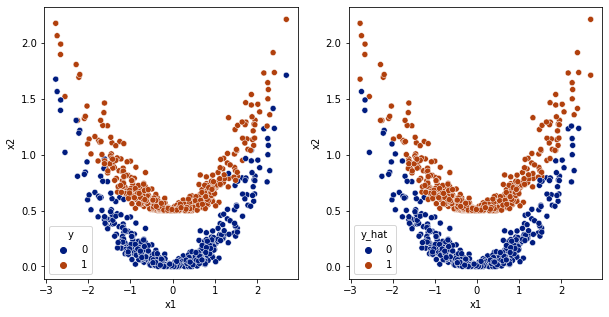
[17]:
y_hat = model.predict(df[['x1','x2']].values)
df['y_hat'] = np.array(y_hat>0.5,dtype='int')
fig,ax = plt.subplots(1,2,figsize=(10,5))
sns.scatterplot(data=df,x='x1',y='x2',hue='y',ax=ax[0], palette='dark')
sns.scatterplot(data=df,x='x1',y='x2',hue='y_hat', ax=ax[1], palette='dark')
[17]:
<AxesSubplot:xlabel='x1', ylabel='x2'>
[18]:
tf.keras.utils.plot_model(model,show_layer_activations=True)
[18]:
[19]:
plot_classification_boundary(model.predict,data=df[['x1','x2','y']].values,size=3,\
figsize=(7,5), bound_details=100)
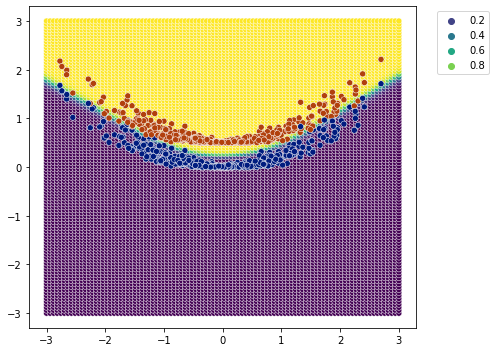
Something with relu and softmax#
for using softmax i’ll have to one hot encode the target.. so in the final layers we can have two outputs
[20]:
from sklearn.preprocessing import OneHotEncoder
[21]:
ohe = OneHotEncoder()
ohe.fit(df[['y']].values)
y_ohe = ohe.transform(df[['y']].values).toarray()
[22]:
model = tf.keras.Sequential([
tf.keras.layers.Dense(units=10, activation='relu'),
tf.keras.layers.Dense(units=10, activation='relu'),
tf.keras.layers.Dense(units=2, activation='softmax')
])
model.compile(
optimizer=tf.optimizers.SGD(learning_rate=0.1),
loss=tf.keras.losses.CategoricalCrossentropy(),
metrics=tf.keras.metrics.CategoricalAccuracy()
)
history = model.fit(
df[['x1','x2']],
y_ohe,
epochs=500,
batch_size=32,
verbose=0,
validation_split = 0.3
)
[23]:
history_metrics = pd.DataFrame(history.history)
history_metrics['epochs'] = history.epoch
fig,ax = plt.subplots(1,2,figsize=(10,5))
history_metrics.plot(x='epochs',y=['loss','val_loss'], ax=ax[0])
history_metrics.plot(x='epochs',y=['categorical_accuracy','val_categorical_accuracy'], ax=ax[1])
[23]:
<AxesSubplot:xlabel='epochs'>
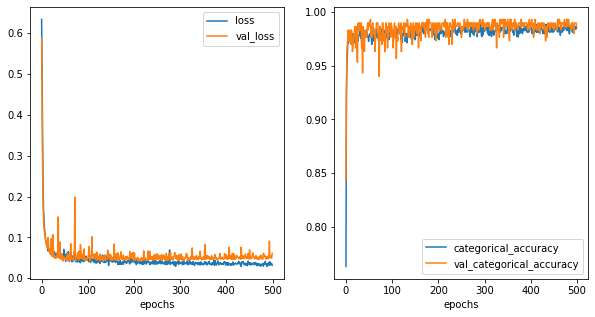
[24]:
y_hat = np.argmax(model.predict(df[['x1','x2']].values),axis=1)
df['y_hat'] = y_hat
fig,ax = plt.subplots(1,2,figsize=(10,5))
sns.scatterplot(data=df,x='x1',y='x2',hue='y',ax=ax[0], palette='dark')
sns.scatterplot(data=df,x='x1',y='x2',hue='y_hat', ax=ax[1], palette='dark')
[24]:
<AxesSubplot:xlabel='x1', ylabel='x2'>
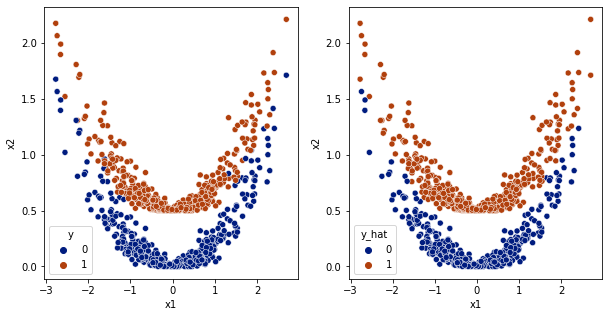
[25]:
tf.keras.utils.plot_model(model,show_layer_activations=True)
[25]:
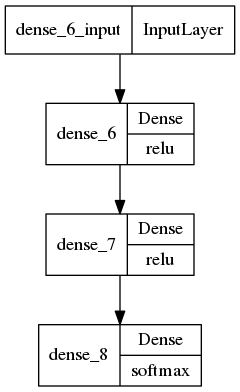
[26]:
plot_classification_boundary(model.predict,data=df[['x1','x2','y']].values,size=3,\
figsize=(10,5), bound_details=100, n_plot_cols=2)
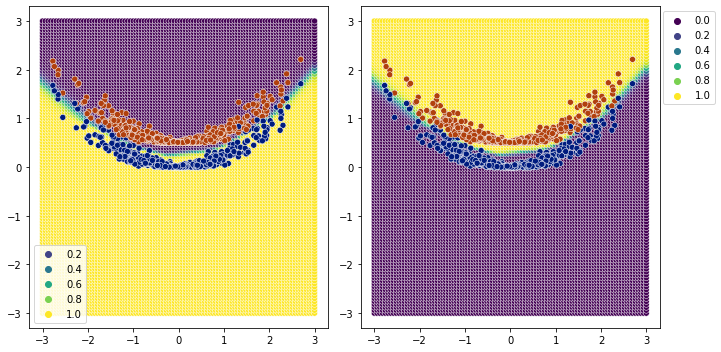
Easy Spiral Classification#
[27]:
from mightypy.ml.dataset import generate_spiral_data
[28]:
data_limit = 30
X, y = generate_spiral_data(data_limit=data_limit, n_classes=3)
[29]:
df = pd.DataFrame(data=X, columns=['x1','x2'])
df['y'] = y
df = df.sample(frac=1)
sns.scatterplot(data=df, x='x1',y='x2',hue='y',palette='dark')
[29]:
<AxesSubplot:xlabel='x1', ylabel='x2'>
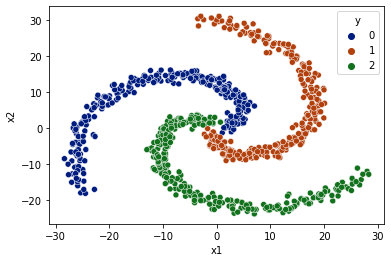
[30]:
ohe = OneHotEncoder()
ohe.fit(df[['y']].values)
y_ohe = ohe.transform(df[['y']].values).toarray()
1 sigmoid#
[31]:
model = tf.keras.Sequential([
tf.keras.layers.Dense(units=64, activation='sigmoid'),
tf.keras.layers.Dense(units=3, activation='softmax')
])
model.compile(
optimizer=tf.optimizers.SGD(learning_rate=1e-2),
loss=tf.keras.losses.CategoricalCrossentropy(),
metrics=tf.keras.metrics.CategoricalAccuracy()
)
history = model.fit(
df[['x1','x2']],
y_ohe,
epochs=500,
batch_size=32,
verbose=0,
validation_split = 0.3
)
history_metrics = pd.DataFrame(history.history)
history_metrics['epochs'] = history.epoch
[32]:
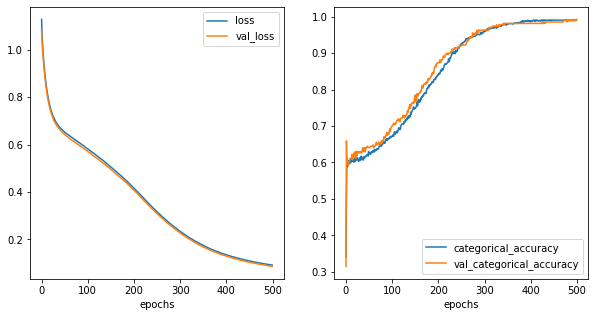
fig,ax = plt.subplots(1,2,figsize=(10,5))
history_metrics.plot(x='epochs',y=['loss','val_loss'], ax=ax[0])
history_metrics.plot(x='epochs',y=['categorical_accuracy','val_categorical_accuracy'], ax=ax[1])
[32]:
<AxesSubplot:xlabel='epochs'>
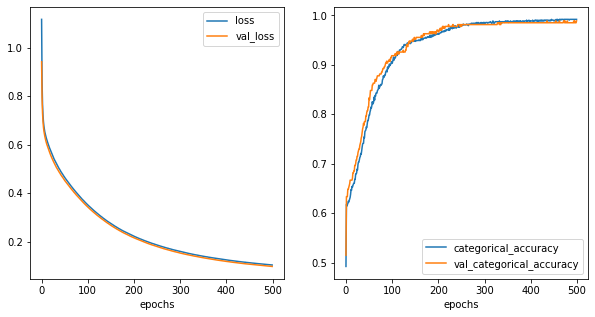
[33]:
df['y_hat'] = np.argmax(model.predict(df[['x1','x2']].values), axis=1)
fig,ax = plt.subplots(1,2,figsize=(10,5))
sns.scatterplot(data=df,x='x1',y='x2',hue='y',ax=ax[0], palette='dark')
sns.scatterplot(data=df,x='x1',y='x2',hue='y_hat', ax=ax[1], palette='dark')
[33]:
<AxesSubplot:xlabel='x1', ylabel='x2'>
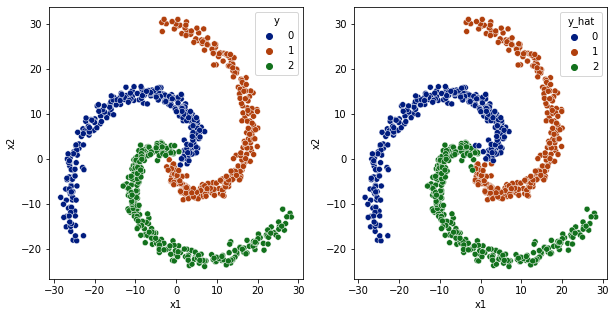
[34]:
plot_classification_boundary(model.predict,data=df[['x1','x2','y']].values,size=30,\
figsize=(15,5), bound_details=100, n_plot_cols=3)
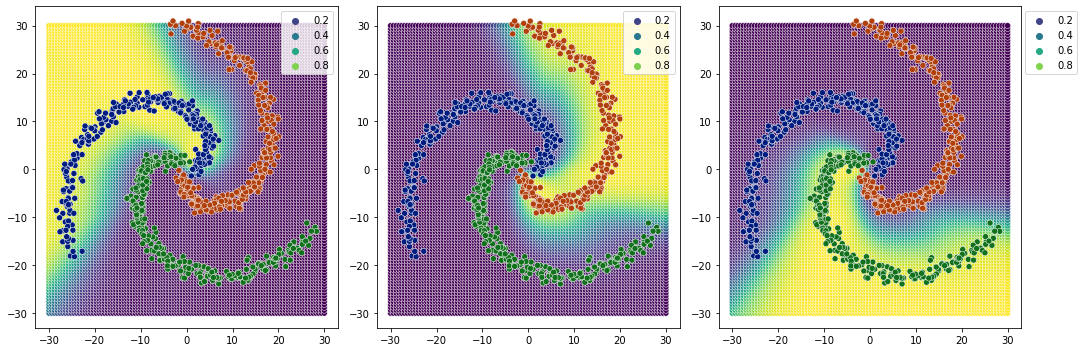
2 sigmoids#
[35]:
model = tf.keras.Sequential([
tf.keras.layers.Dense(units=64, activation='sigmoid'),
tf.keras.layers.Dense(units=64, activation='sigmoid'),
tf.keras.layers.Dense(units=3, activation='softmax')
])
model.compile(
optimizer=tf.optimizers.SGD(learning_rate=1e-2),
loss=tf.keras.losses.CategoricalCrossentropy(),
metrics=tf.keras.metrics.CategoricalAccuracy()
)
history = model.fit(
df[['x1','x2']],
y_ohe,
epochs=500,
batch_size=32,
verbose=0,
validation_split = 0.3
)
history_metrics = pd.DataFrame(history.history)
history_metrics['epochs'] = history.epoch
[36]:
fig,ax = plt.subplots(1,2,figsize=(10,5))
history_metrics.plot(x='epochs',y=['loss','val_loss'], ax=ax[0])
history_metrics.plot(x='epochs',y=['categorical_accuracy','val_categorical_accuracy'], ax=ax[1])
[36]:
<AxesSubplot:xlabel='epochs'>
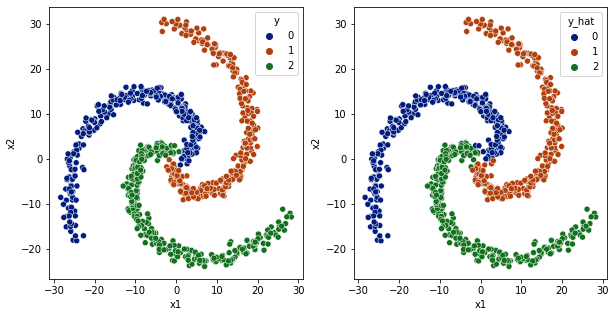
[37]:
df['y_hat'] = np.argmax(model.predict(df[['x1','x2']].values), axis=1)
fig,ax = plt.subplots(1,2,figsize=(10,5))
sns.scatterplot(data=df,x='x1',y='x2',hue='y',ax=ax[0], palette='dark')
sns.scatterplot(data=df,x='x1',y='x2',hue='y_hat', ax=ax[1], palette='dark')
[37]:
<AxesSubplot:xlabel='x1', ylabel='x2'>
[38]:
plot_classification_boundary(model.predict,data=df[['x1','x2','y']].values,size=30,\
figsize=(15,5), bound_details=100, n_plot_cols=3)
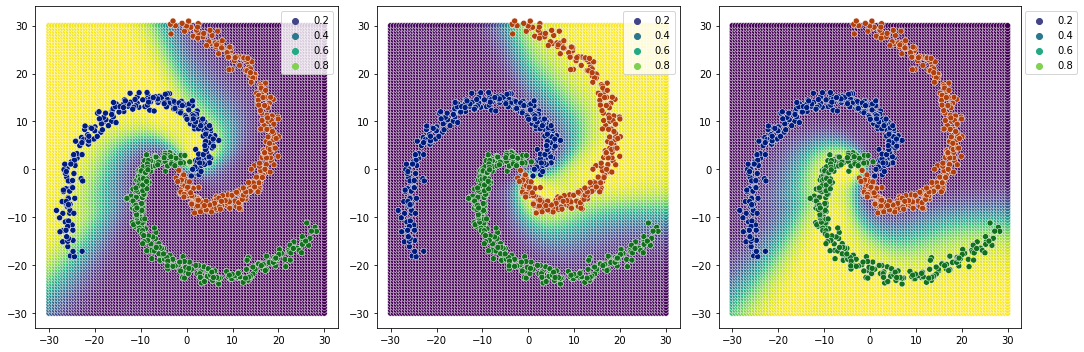
1 relu layer#
[39]:
model = tf.keras.Sequential([
tf.keras.layers.Dense(units=64, activation='relu'),
tf.keras.layers.Dense(units=3, activation='softmax')
])
model.compile(
optimizer=tf.optimizers.Adam(learning_rate=1e-2),
loss=tf.keras.losses.CategoricalCrossentropy(),
metrics=tf.keras.metrics.CategoricalAccuracy()
)
history = model.fit(
df[['x1','x2']],
y_ohe,
epochs=500,
batch_size=32,
verbose=0,
validation_split = 0.3
)
history_metrics = pd.DataFrame(history.history)
history_metrics['epochs'] = history.epoch
[40]:
fig,ax = plt.subplots(1,2,figsize=(10,5))
history_metrics.plot(x='epochs',y=['loss','val_loss'], ax=ax[0])
history_metrics.plot(x='epochs',y=['categorical_accuracy','val_categorical_accuracy'], ax=ax[1])
[40]:
<AxesSubplot:xlabel='epochs'>
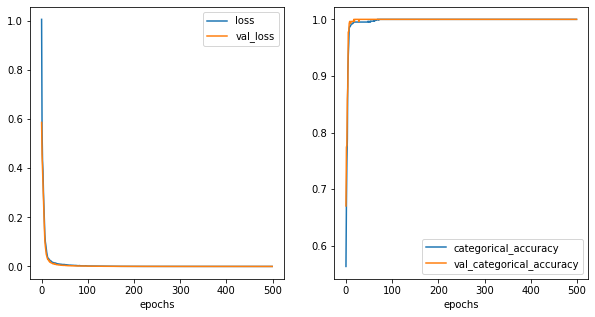
[41]:
df['y_hat'] = np.argmax(model.predict(df[['x1','x2']].values), axis=1)
fig,ax = plt.subplots(1,2,figsize=(10,5))
sns.scatterplot(data=df,x='x1',y='x2',hue='y',ax=ax[0], palette='dark')
sns.scatterplot(data=df,x='x1',y='x2',hue='y_hat', ax=ax[1], palette='dark')
plt.show()
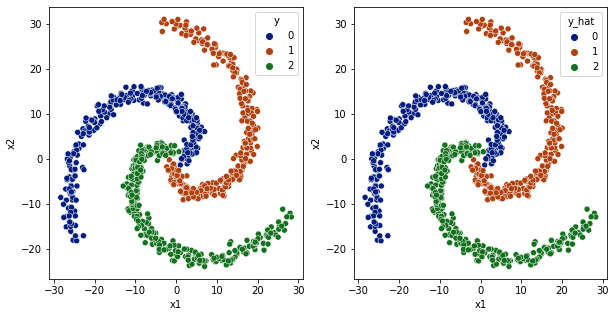
[42]:
plot_classification_boundary(model.predict,data=df[['x1','x2','y']].values,size=30,\
figsize=(15,5), bound_details=100, n_plot_cols=3)
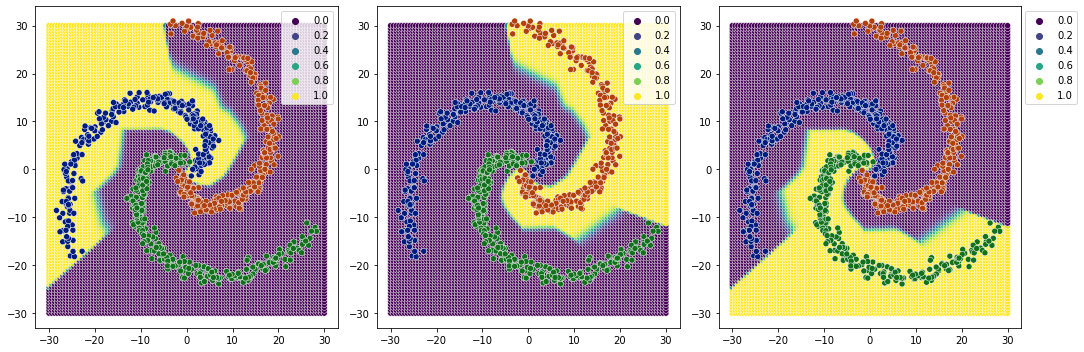
relu as compared to sigmoid, creates sharper edges(more clear probabilities).
2 relu layers#
[43]:
model = tf.keras.Sequential([
tf.keras.layers.Dense(units=100, activation='relu'),
tf.keras.layers.Dense(units=100, activation='relu'),
tf.keras.layers.Dense(units=3, activation='softmax')
])
model.compile(
optimizer=tf.optimizers.Adam(learning_rate=0.01),
loss=tf.keras.losses.CategoricalCrossentropy(),
metrics=tf.keras.metrics.CategoricalAccuracy()
)
history = model.fit(
df[['x1','x2']],
y_ohe,
epochs=500,
batch_size=32,
verbose=0,
validation_split = 0.3
)
history_metrics = pd.DataFrame(history.history)
history_metrics['epochs'] = history.epoch
[44]:
fig,ax = plt.subplots(1,2,figsize=(10,5))
history_metrics.plot(x='epochs',y=['loss','val_loss'], ax=ax[0])
history_metrics.plot(x='epochs',y=['categorical_accuracy','val_categorical_accuracy'], ax=ax[1])
[44]:
<AxesSubplot:xlabel='epochs'>
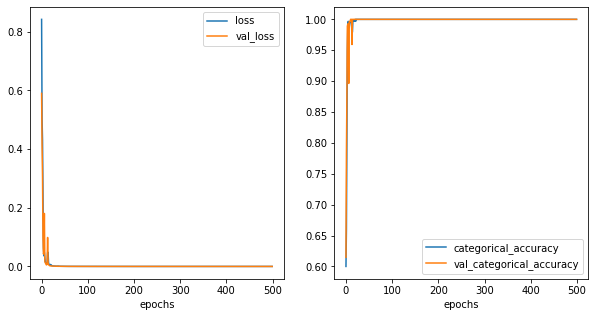
[45]:
df['y_hat'] = np.argmax(model.predict(df[['x1','x2']].values), axis=1)
fig,ax = plt.subplots(1,2,figsize=(10,5))
sns.scatterplot(data=df,x='x1',y='x2',hue='y',ax=ax[0], palette='dark')
sns.scatterplot(data=df,x='x1',y='x2',hue='y_hat', ax=ax[1], palette='dark')
plt.show()
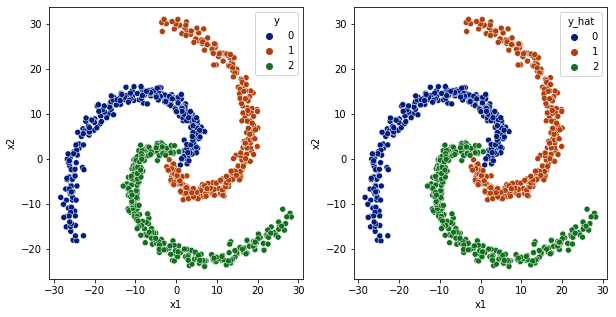
[46]:
plot_classification_boundary(model.predict,data=df[['x1','x2','y']].values,size=30,\
figsize=(15,5), bound_details=100, n_plot_cols=3)
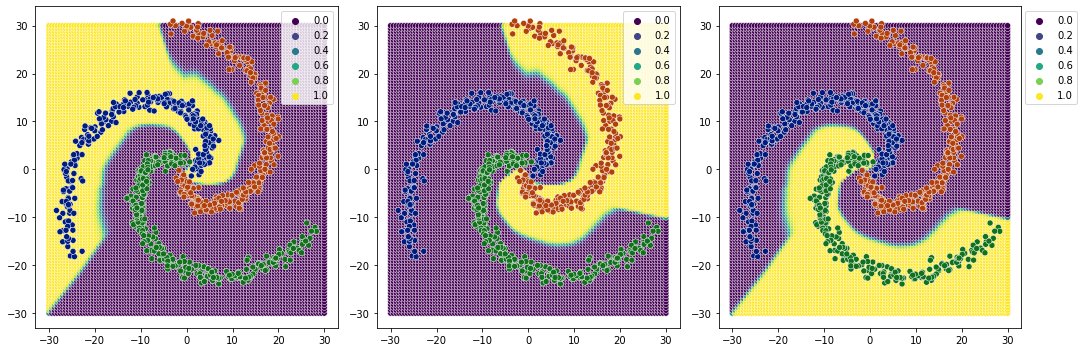
1 layer relu almost did the job. 2 layer relu is almost the same.
Complex Spiral Classification#
[47]:
data_limit = 100
X, y = generate_spiral_data(data_limit=data_limit, n_classes=3)
df = pd.DataFrame(data=X, columns=['x1','x2'])
df['y'] = y
df = df.sample(frac=1)
sns.scatterplot(data=df, x='x1',y='x2',hue='y',palette='dark')
[47]:
<AxesSubplot:xlabel='x1', ylabel='x2'>
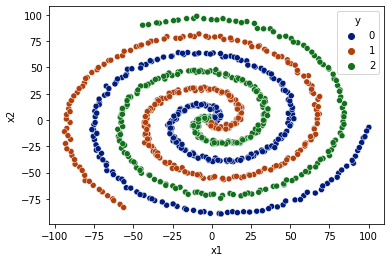
[48]:
ohe = OneHotEncoder()
ohe.fit(df[['y']].values)
y_ohe = ohe.transform(df[['y']].values).toarray()
1 relu layer#
[49]:
model = tf.keras.Sequential([
tf.keras.layers.Dense(units=100, activation='relu'),
tf.keras.layers.Dense(units=3, activation='softmax')
])
model.compile(
optimizer=tf.optimizers.Adam(learning_rate=0.01),
loss=tf.keras.losses.CategoricalCrossentropy(),
metrics=tf.keras.metrics.CategoricalAccuracy()
)
history = model.fit(
df[['x1','x2']],
y_ohe,
epochs=500,
batch_size=32,
verbose=0,
validation_split = 0.2
)
history_metrics = pd.DataFrame(history.history)
history_metrics['epochs'] = history.epoch
[50]:
fig,ax = plt.subplots(1,2,figsize=(10,5))
history_metrics.plot(x='epochs',y=['loss','val_loss'], ax=ax[0])
history_metrics.plot(x='epochs',y=['categorical_accuracy','val_categorical_accuracy'], ax=ax[1])
[50]:
<AxesSubplot:xlabel='epochs'>
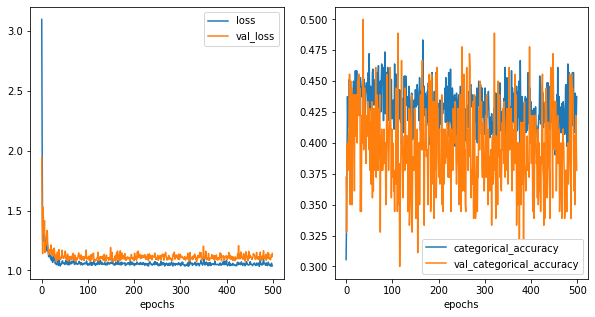
[51]:
df['y_hat'] = np.argmax(model.predict(df[['x1','x2']].values), axis=1)
fig,ax = plt.subplots(1,2,figsize=(10,5))
sns.scatterplot(data=df,x='x1',y='x2',hue='y',ax=ax[0], palette='dark')
sns.scatterplot(data=df,x='x1',y='x2',hue='y_hat', ax=ax[1], palette='dark')
plt.show()
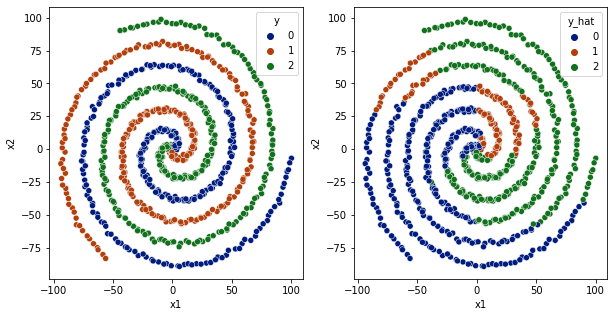
[52]:
plot_classification_boundary(model.predict,data=df[['x1','x2','y']].values,size=data_limit,\
figsize=(15,5), bound_details=100, n_plot_cols=3)
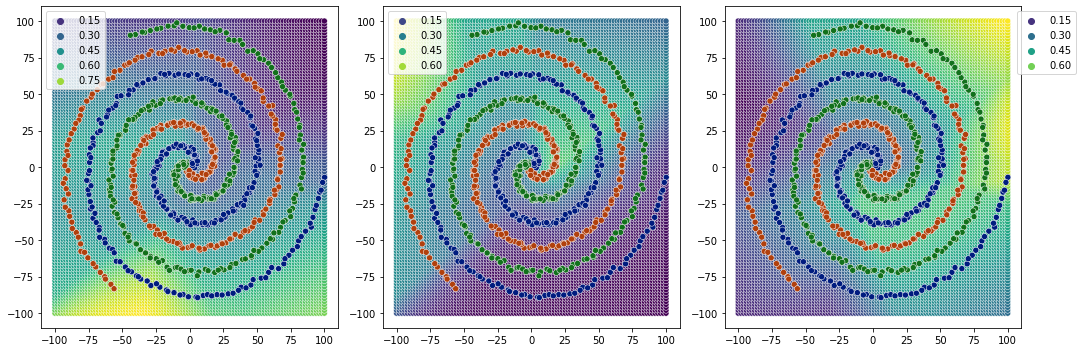
So, for 1 relu layer, it is not able to converge. and decision boundaries are not very clear.
2 relu layers#
[53]:
model = tf.keras.Sequential([
tf.keras.layers.Dense(units=100, activation='relu'),
tf.keras.layers.Dense(units=100, activation='relu'),
tf.keras.layers.Dense(units=3, activation='softmax')
])
model.compile(
optimizer=tf.optimizers.Adam(learning_rate=0.001),
loss=tf.keras.losses.CategoricalCrossentropy(),
metrics=tf.keras.metrics.CategoricalAccuracy()
)
history = model.fit(
df[['x1','x2']],
y_ohe,
epochs=500,
batch_size=32,
verbose=0,
validation_split = 0.3
)
history_metrics = pd.DataFrame(history.history)
history_metrics['epochs'] = history.epoch
[54]:
fig,ax = plt.subplots(1,2,figsize=(10,5))
history_metrics.plot(x='epochs',y=['loss','val_loss'], ax=ax[0])
history_metrics.plot(x='epochs',y=['categorical_accuracy','val_categorical_accuracy'], ax=ax[1])
[54]:
<AxesSubplot:xlabel='epochs'>
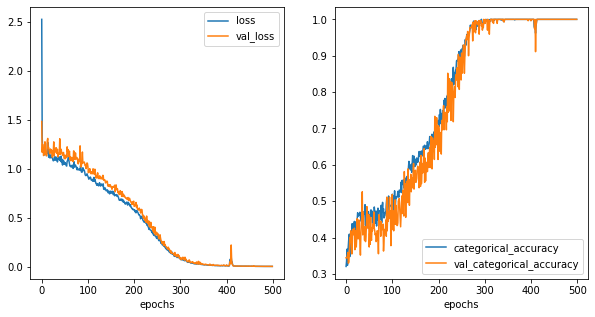
[55]:
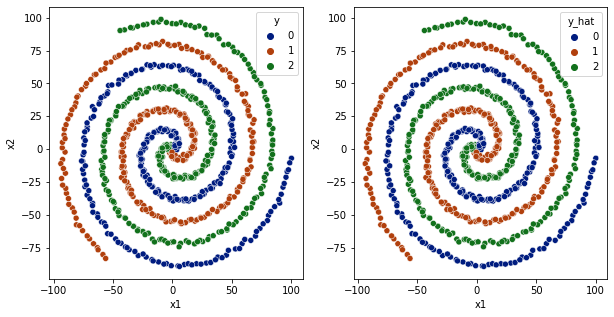
df['y_hat'] = np.argmax(model.predict(df[['x1','x2']].values), axis=1)
fig,ax = plt.subplots(1,2,figsize=(10,5))
sns.scatterplot(data=df,x='x1',y='x2',hue='y',ax=ax[0], palette='dark')
sns.scatterplot(data=df,x='x1',y='x2',hue='y_hat', ax=ax[1], palette='dark')
plt.show()
[56]:
plot_classification_boundary(model.predict,data=df[['x1','x2','y']].values,size=data_limit,\
figsize=(15,5), bound_details=100, n_plot_cols=3)
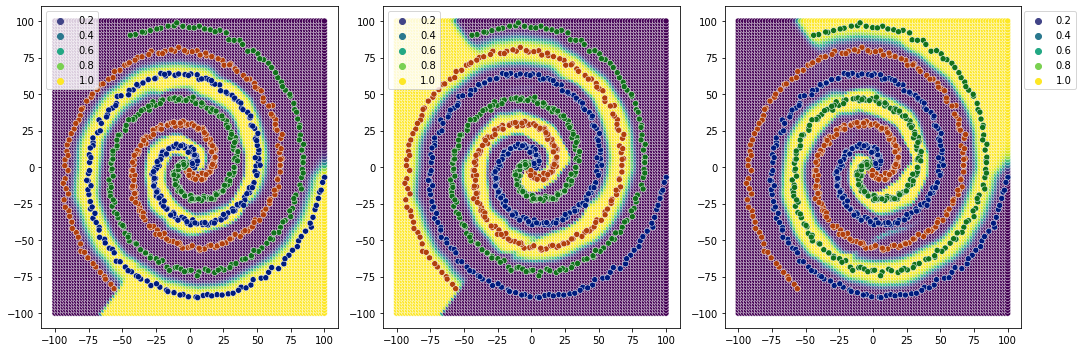
Clear decision boundaries.
A little bit complex model#
[57]:
model = tf.keras.Sequential([
tf.keras.layers.Dense(units=100, activation='relu'),
tf.keras.layers.Dense(units=100, activation='relu'),
tf.keras.layers.Dense(units=100, activation='relu'),
tf.keras.layers.Dense(units=100, activation='relu'),
tf.keras.layers.Dense(units=3, activation='softmax')
])
model.compile(
optimizer=tf.optimizers.Adam(learning_rate=0.001),
loss=tf.keras.losses.CategoricalCrossentropy(),
metrics=tf.keras.metrics.CategoricalAccuracy()
)
history = model.fit(
df[['x1','x2']],
y_ohe,
epochs=500,
batch_size=32,
verbose=0,
validation_split = 0.3
)
history_metrics = pd.DataFrame(history.history)
history_metrics['epochs'] = history.epoch
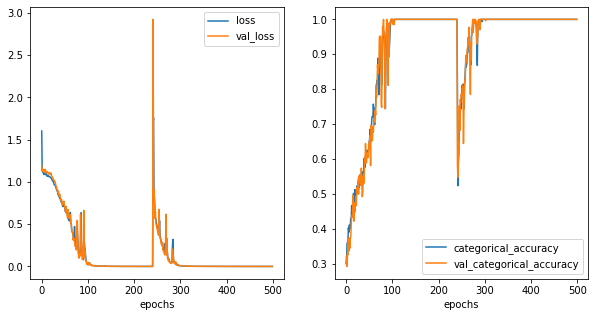
[58]:
fig,ax = plt.subplots(1,2,figsize=(10,5))
history_metrics.plot(x='epochs',y=['loss','val_loss'], ax=ax[0])
history_metrics.plot(x='epochs',y=['categorical_accuracy','val_categorical_accuracy'], ax=ax[1])
[58]:
<AxesSubplot:xlabel='epochs'>
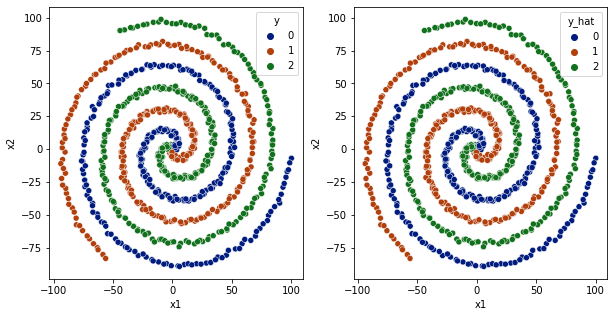
[59]:
df['y_hat'] = np.argmax(model.predict(df[['x1','x2']].values), axis=1)
fig,ax = plt.subplots(1,2,figsize=(10,5))
sns.scatterplot(data=df,x='x1',y='x2',hue='y',ax=ax[0], palette='dark')
sns.scatterplot(data=df,x='x1',y='x2',hue='y_hat', ax=ax[1], palette='dark')
plt.show()
[60]:
plot_classification_boundary(model.predict,data=df[['x1','x2','y']].values,size=data_limit,\
figsize=(15,5), bound_details=100, n_plot_cols=3)
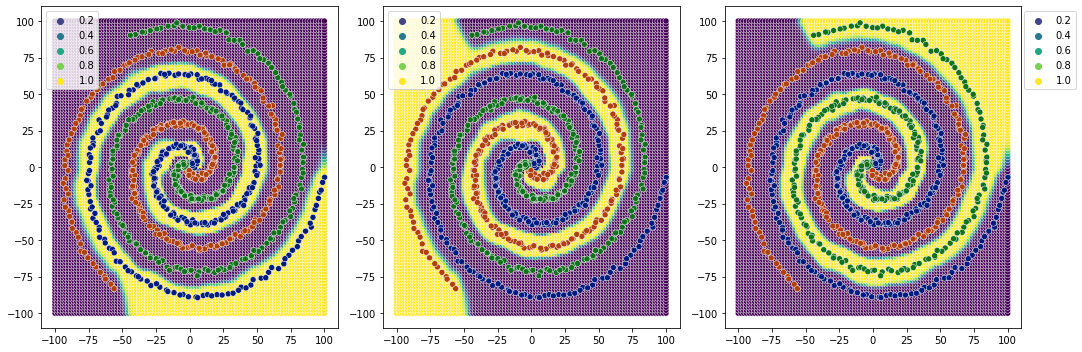
clearer decision boundaries than 2 relu layers.